OAI Analysis 2
This software contains open-source analysis approaches for the Osteoarthritis Initiative (OAI) magnetic resonance image (MRI) data. The analysis code is largely written in Python with the help of ITK and VTK for data I/O and mesh processing as well as PyTorch for the deep learning approaches for segmentation and registration.
The following functionality is currently supported:
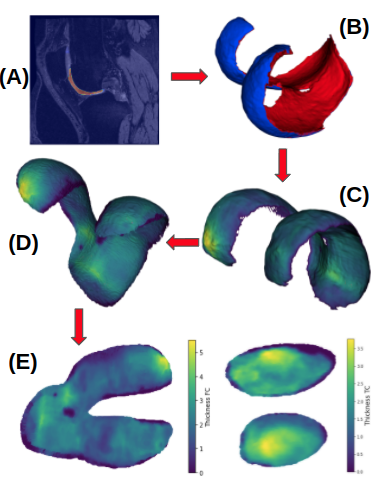
- Deep Learning Segmentation: Automatic cartilage segmentation (femoral and tibial cartilage) using a 3D UNet.
- Cartilage thickness: Extraction of femoral and tibial cartilage meshes and measuring of cartilage thickness based on a closest-point thickness estimation.
- Deep Learning Atlas Registration: Registration of the carilage meshes with associated cartilage thickness to a knee atlas space via a deep registration network.
- 2D Thickness Mapping: Mapping of the thickness maps to a common 2D atlas space which provides full spatial correspondence. This is achieved via unrolling (based on a cylindrical coordinate system) for the femoral cartilage and a planar projection for the tibial cartilage.
- Statistical Analysis: [Longitudinal statistical analysis approaches will be added shortly]
The OAI Analysis 2 repository can be found here: https://github.com/uncbiag/OAI_analysis_2