Osteoarthritis
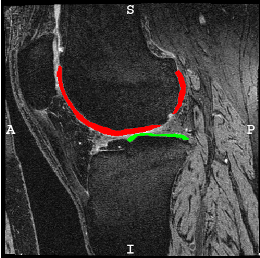
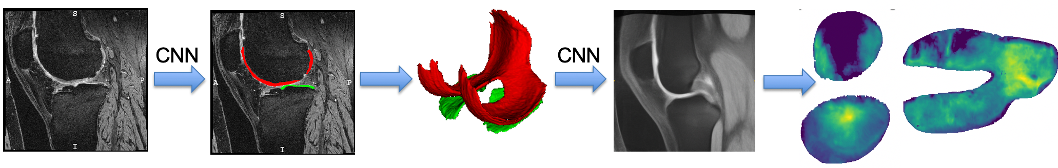
The goal of this project is to optimize and openly provide to the osteoarthritis (OA) community a new technology to rapidly and automatically measure cartilage thickness, appearance and changes on magnetic resonance images (MRI) of the knee for very large image databases. This will allow prediction of OA progression and associations of risk factors for OA with OA progression on an unprecedented scale; future work will focus on incorporating additional disease markers, ranging from MRI-derived biomarkers for bone and synovial lesions, to biochemical biomarkers, to genetic information. OA is the most frequent form of arthritis and a common cause of disability. Knee OA (KOA), in particular, leads to substantial morbidity due to mobility limitations and is the most common indication for total knee arthroplasty (TKA). Although OA is a disease of the joint as an organ, cartilage loss is a central feature.
MRI is a promising technology for more detailed and sensitive assessments of KOA for research purposes. However, MRI technology remains expensive, and although acquisition times have gradually decreased, the post-processing, segmentation, and interpretation of the images remain a rate-limiting step, particularly for large cohort studies. Hence, large datasets cannot be meaningfully analyzed, resulting in multiple small, under-powered studies of select groups. This particularly limits studies of infrequent outcomes (e.g. TKA, disability). Furthermore, the lack of sensitive outcome measures for OA progression limits the ability to establish associations with putative OA risk factors.
Our group initially developed a reliable, validated, and automated atlas-based cartilage segmentation method that allows rapid determination of local cartilage thickness over a large number of images [shan14] . This method successfully analyzed the full dataset for the Pfizer Longitudinal Study (>700 images total) in about 10,000 CPU hours (four days on a medium-sized cluster) and allowed assessment of cartilage thickness across all time points and all available images in the cohort. We are currently working on advanced analysis approaches that are applicable to datasets orders of magnitude larger. Specifically our goal is the analysis of the full image dataset of the Osteoarthritis Initiative (OAI). So far we have developed a variety of approaches for automatic cartilage segmentation ( [xu18] [xu19] ) and registration via deep learning ( [shen19a] [shen19b] ). We have also developed approaches to quantify cartilage thickness based on the resulting cartilage segmentations and to evaluate them in a common atlas coordinate system. Using these approaches, we have successfully processed the entire OAI image dataset (on the order of 40,000 3D images). We have also developed various longitudinal statistical analysis approachs (see for example, [huang15] ) and are currently working on applying extensions of these approaches to the OAI data. Lately, we have also focused efforts on data augmentation approaches which allow training deep segmentation networks with small numbers of labeled images ( [shen20] [olut20] ).
Acknowledgements
This work is currently supported by the National Institutes of Health (NIH) under award number NIH 1R01AR072013. The content of our work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
| shan14 |
| huang15 |
| shen19a |
| shen19b |
| xu18 |
| xu19 |